ANNE: A weight engineering approach that extracts encoded information in an artificial neural network model via manipulating neuronal weights learned from data | |||||
|
|||||
Summary ^
Artificial Neural Network Encoder (ANNE), a novel weight engineering method
developed in Hu Li's lab that harness the
power of autoencoder and demonstrated that it is possible to decode
meaningful information encoded in ANN models trained for specific tasks.
We applied ANNE on breast cancer gene expression data with known clinical
properties as case studies. Our work illustrates the trained autoencoder
models are indeed information encoders that meaningful gene-gene
associations with numerous supported evidences can be retrieved. This work
therefore opens a new avenue in machine intelligence that ANN models will
no longer perceived as tools to perform recognition tasks but as powerful
tools to extract meaningful information embedded within the sea of high
dimensional data.
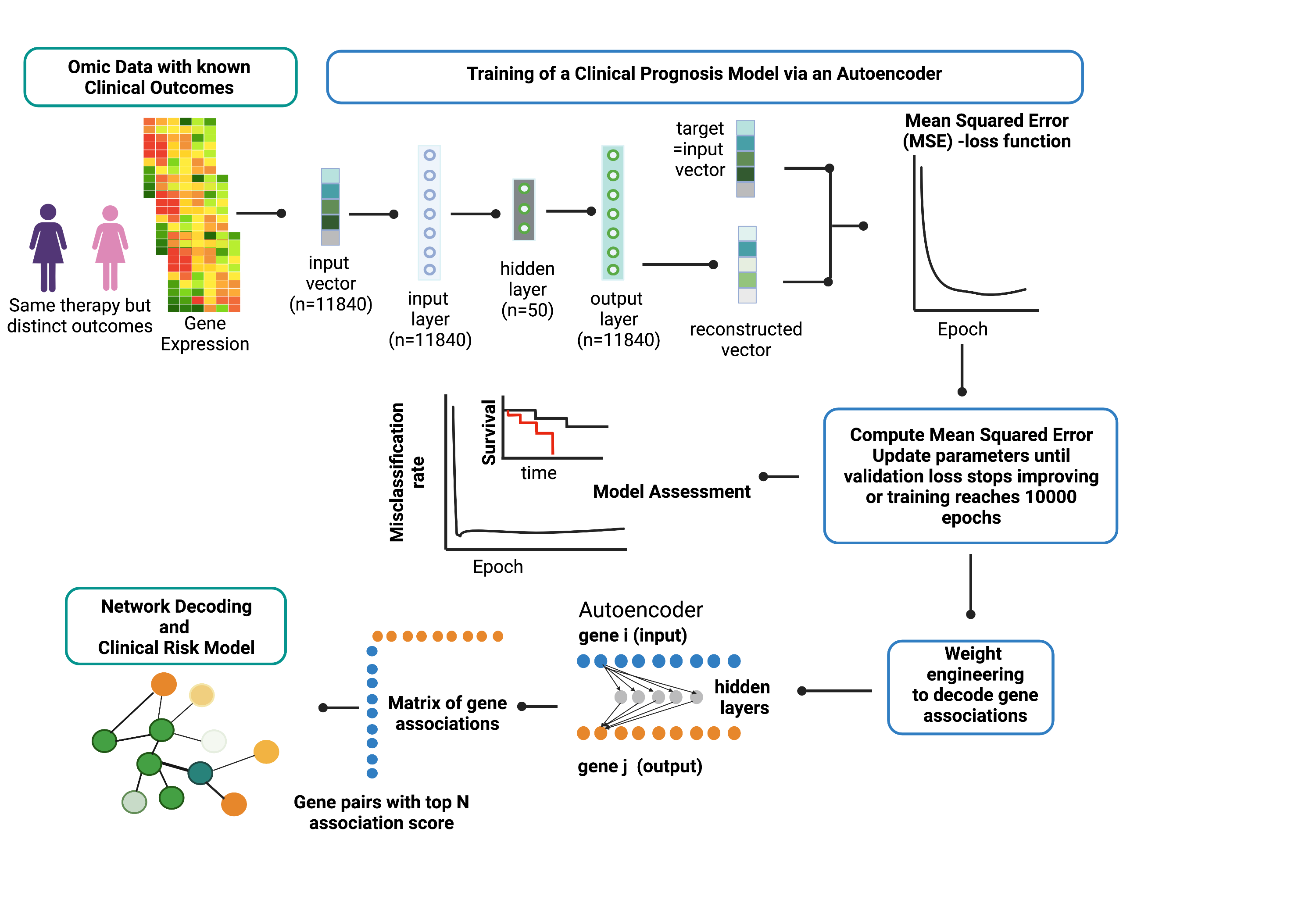
Overview of Artificial Neural Network Encoder (ANNE) algorithm using breast cancer prognosis as an illustrative example. The ANNE algorithm uses gene expression profiles from breast cancer patients with known prognostic outcomes to simultaneously decode gene-gene associations, networks and derive clinical risk models. Patient samples are assigned to either good or poor prognosis according to disease relapse-free survival (DRFS). Next, an autoencoder algorithm is used to train the models, with an input vector that represents all genes (features) present in transcriptomics data and each gene corresponds to a node in the input layer. The dimensionality of the output layer (i.e., number of nodes) is the same as the input layer. The autoencoder has an architecture of one input layer, one hidden layer and one output layer. During the training phase the autoencoder reconstruct values from input layer into an output layer and the output vector is compared with input vector to compute the reconstruction error. The training process is repeated by updating weights connecting nodes (or neurons) from input layer to hidden layer and from hidden layer to output layer via a backpropagation algorithm. The training process is repeated by feeding input through the neural network to the output layer to calculate the training loss, and updating the weights connecting nodes from the output layer to hidden layer and to the input layer via a backpropagation algorithm. Training process is complete when no further improvements on the reconstruction error is achieved, or training has reached certain number epochs. Next, the learned weights connecting all nodes from input to output layers in a trained autoencoder model are used to decode meaningful gene-gene associations using an association scoring scheme. Computed association scores for all gene pairs are aggregated to an association score matrix and the top gene pairs (n=200) with highest absolute scores are selected to build ANNE-decoded networks. Genes that occur multiple times will serve as "anchors" to agglomerate gene pairs into a network. Figure gen Results ^
Explore gene association networks derived from autoencoder models for phenotype groups here. (Please view the link in the Chrome or Firefox web browser.) Download ^
Download the scripts on GitHub
with sample dataset to run ANNE on your local Linux system.
Extract the scripts and sample datasets into the same folder
and read the README file to get started. Support ^
For support or questions of ANNE, please post to our google group. Citation ^
Manuscript in preparation. |